Introduction¶
DCEST/COS-CEST experiments¶. Nmrglue.agilent; Relaxation experiments¶. Se details at: relax with OpenDX. NMRPipe. MddNMR. nmrglue. Art Palmers software: ModelFree4.
nmrglue is a python module for reading, writing, and interacting with thespectral data stored in a number of common NMR data formats. This tutorialprovides an overview of some of the features of nmrglue. A basicunderstanding of python is assumed which can be obtained by reading someof the python documentation. The examples inthis tutorial can be run interactively from the python shell but the use of anenhanced python shell which provides non-blocking control of GUI threads,for example ipython, isrecommended when trying the examples which use matplotlib. The sample datausing in this tutorial isavailable is you wish tofollow along with the same files.
The software can also be used in Google Colabs - see details below.
Reading NMR files¶
nmrglue can read and write to a number of common NMR file formats. To seehow simple this can be let’s read a 2D NMRPipe file. (Note: If you need an exampledataset, use the one provided here, and replace test.fid or test.ft with the namesof the files ending in .fid or .dat).
Here we have imported the nmrglue module and opened the NMRPipe filetest.fid. nmrglue contains a number of modules for reading and writing NMRfiles and all of these modules have a read function which opens a fileor directory containing NMR data, reads in any necessary information, and loadsthe spectral data into memory. The read function returns a 2-tuplecontaining a python dictionary with file and spectral parameters and anumpy array object containing the numericspectral data. Currently the following file formats are supported by nmrgluewith the associated module:
Module | File Format | Reference |
|---|---|---|
bruker | Bruker | |
pipe | NMRPipe | |
sparky | Sparky | |
varian | Varian/Agilent | |
rnmrtk | Rowland NMR Toolkit | |
jcampdx | JCAMP-DX | |
nmrml | NMR Markup Language | |
simpson | Simpson | |
tecmag | Technology for MagneticResonance |
Examining the data object in more detail:
We can see that this is a two dimensional data set with 1500 complex pointsin the direct dimension and 332 points in the indirect dimension. nmrgluetakes care of converting the raw data in the file into an array of appropriatetype, dimensionality, and quadrature. For complex data the last axis,typically the direct dimension, is converted to a complex data type. The otheraxes are not converted.
In some cases, not all of the information needed to represent the spectral dataas a well formed numpy array is stored in the file, or the values determinedautomatically are incorrect. In many of these cases, this information can bespecified directly in the function call.
For example, the read function in the varian module sometimes cannotdetermine the shape or fid ordering of 3D files correctly. These parameterscan be explicitly provided in the function call with the shape and torderkeywords. See nmrglue.varian for details.
Universal dictionaries¶
In addition to the spectral data, the read function also determinesvarious spectral parameters that were stored in the file and stores them in apython dictionary:
Here we see NMRPipe files stores the spectal width of the direct dimension(50000.0 Hz) and the name of the indirect dimension (15N) as well as a numberof additional parameters.
Some file formats describe well the spectral data, listing a large number ofparameters, other only a few. In addition, different formats expressparameters in different units and under different names. For users who arefamiliar with the specific file format or are working with only a single filetype, this is not a problem; the dictionary allows direct access to theseparameters. If a more uniform listing of spectral parameters is desired, theguess_udic function can be used to create a ‘universal’ dictionary.
This ‘universal’ dictionary of spectral parameters contains only the mostfundamental parameters, the dimensionality of the data, and a dictionaryof parameters for each axis numbered according to the data array ordering(the direct dimension is the highest numbered dimension). The axisdictionaries contain the following keys:
Key | Description |
|---|---|
car | Carrier frequency in Hz. |
complex | True for complex data, False for magnitude data. |
encoding | How the data is encoded, ‘states’, ‘tppi’, etc. |
freq | True for frequency domain data, False for time domain. |
label | String describing the axis name. |
obs | Observation frequency in MHz. |
size | Dimension size (R|I for last axis, R+I for others) |
sw | Spectral width in Hz. |
time | True for time domain data, False got frequency domain. |
For our 2D NMRPipe file, these parameters for the indirect dimension are:
One note on the size key, it was designed to always match the shape of thedata:
Not all NMR files formats contain all the information necessary to determineuniquely all of the universal dictionary parameters. In these cases, thedictionary will be filled with generic values (999.99, “X”, “Y”, etc) andshould be updated by the user with the correct values.In converting to a ‘universal’ dictionary we have sacrificed additionalinformation about the data which was contained in the original file in orderto provide a common description of NMR data. Despite the universaldictionary’s limited information, together with the data array, it is sufficientfor most NMR tasks. We will later see that the universal dictionary allowsfor conversions between file formats.
Manipulating NMR data¶
Let us return again to the data array. By providing direct access to thespectral data as a numpy array we can examine and manipulate this data usinga number of simple methods as well as a number of functions. Sincethe read function moves the data into memory all this data manipulationis done without effecting the original data file.
We can use slices to examine single values in the array:
Or an whole vector:
And along the indirect dimension:
We can do more advanced slicing:
If we just want the real or imaginary channel:
We find characteristics of the data:
Reshape or transpose the data:
Finally we can set the value of data as desired. For example setting asingle point:
Or a region:
The numpy documentation has additionalinformation on thearrayobject. In addition by combining nmrglue withnumpy and/or scipymore complex data manipulation and calculation can be performed. Later wewill show how these modules are used to create a full suite of processingfunctions.
Writing NMR files¶
Now that we have modified the original NMR data we can write our modificationto a file. nmrglue again makes this simple:
Reading in both the original data and this new data we can see that they aredifferent:
The parameter dictionary has not changed:
By default nmrglue will not overwrite existing data with the writefunction:
But this check can be by-passed with the overwrite parameter:
The unit_conversion object¶
Earlier we used the array index values for slicing the numpy array. Forreference your data in more common NMR units nmrglue provides theunit_coversion object. Use the make_uc function to create aunit_conversion object:

We now have unit conversion objects for both axes in the 2D spectrum. We canuse these objects to determined the nearest point for a given unit:
Or an exact value:
We can also convert from points to various units:
These objects can also be used for slicing, for example to find the tracecloses to 120 ppm:
Converting between file formats¶
nmrglue can also be used to convert between file formats using the convertmodule. For example to convert a 2D NMRPipe file to a Sparky file:
Nmrglue Examples
Here we opened the NMRPipe file test.ft2 , created a new converter objectand loaded it with the NMRPipe data. The converter is then used to generatethe Sparky parameter dictionary and a data array appropriate for Sparky datawhich is written to sparky_file.ucsf.All type conversions, and sign manipulation of the data array is performedinternally by the converter object. In addition new dictionaries arecreated from an internal universal dictionary for the desired output.Additional examples showing how to use nmrglue to convert between NMR fileformats can be found in the Convert Examples.
Low memory reading/writing of files¶
Up to this point we have read NMR data from files using the read function.This function reads the spectral data from a NMR file into the computersmemory. For small data sets this is fine, modern computer have sufficientRAM to store complete 1D and 2D NMR data sets and a few copies of thedata while processing. For 3D and larger dimensionality data set this is oftennot desired. Reading in an entire 3D data set is not required when only asmall portion must be examined for viewing or processing. With this in mindnmrglue provides methods to read only a portions of NMR data from files whenit is required. This is accomplished by creating a new object which lookvery similar to numpy array but does not load data into memory.Rather when a particular slice is requested the object opens thenecessary file(s), reads in the data and returns to the user a numpyarray with the data. In addition these objects have transpose and swapaxesmethod and can be iterated over just as numpy arrays but without usinglarge amounts of memory. The only limitation of these objects is that theydo not support assignment, so a slice must be taken before changing the valueof data. The fileio sub-modules all have some form of read_lowmemfunction which return these low-memory objects. For example reading the 2Dsparky file we created earlier:
Slicing returns a numpy array:
The data can be transposed as a numpy array:
These low memory usage objects can be written to disk or used in toload a conversion object just as if they were normal numpy arrays.
Similar when large data sets are to be written to disk, it often doesnot make sense to write the entire data set at once. For this thewrite_lowmem functions in the fileIO submodules provide methods fortrace-by-trace or similar writing.
Processing data¶
With NMR spectral data being stored as a numpy array a number of linearalgebra and signal processing functions can be applied to the data. Thefunctions in the numpyand scipy modules offer a number of processingfunctions users might find useful. nmrglue provides a number of commonNMR functions in the nmrglue.proc_base module, baseline related functionsin nmrglue.proc_bl, and linear prediction functions in the nmrglue.proc_lpmodule. For example we perform some simple processing on our 2D NMRPipe file(output suppressed):
These functions process only the data, they do notupdate the spectral parameter associated with the data. Because thesevalues are key when examining NMR data we want functions which take intoaccount these parameter while processing. nmrglue provides thenmrglue.pipe_proc module for processing NMRPipe data while updating thespectral properties simultaneously. Additional modules for processingother file format are being developed. Using pipe_proc is similar tousing NMRPipe itself. For example to process the sample 2D NMRPipe file:
This processed file can then be written out
In the example above the entire data set was processed in memory. All theprocessing functions were applied to a set of data stored in the computersRAM after which the entire 2D data set was written to disk. For 1D and 2Ddata sets this is fine, but as mentioned earlier many 3D and larger data setscannot be processed in this manner. For a 3D file what is desired is thateach 2D XY plane be read, processed and saved. Then the ZX planes are readfrom this new file, the Z plane processed and these planes saved into thefinal file. In nmrglue this can be accomplished for NMRPipe files using theiter3D object. Currently no other file format allowssuch processing but development of these is planned.An example of processing a 3D NMRPipe file using a iter3D object can befound in process example: process_pipe_3d.
Additional examples showing how to use nmrglue to process NMR data can befound in the Processing Examples.
Using matplotlib to create figures¶
A number of python plotting libraries exist which can be used in conjunctionwith nmrglue to produce publication quality figures. matplotlib is one ofthe more popular libraries and has the ability to output to a number ofhard copy formats as well as offering a robust interactive environment. Whenusing matplotlib interactively use of ipythonor a similar shell is recommended although the standard python shell can beused.
Here we have loaded the pyplot module from matplotlib (aliased as plt), andused it to plot the 1D frequency domain data of a model protein. The resultingfigure is saved as plot_1d.png.
Alternately, the object-oriented interface from matplotlib can be used. This is especially useful when make more complicated plots. The above example would look something like this:
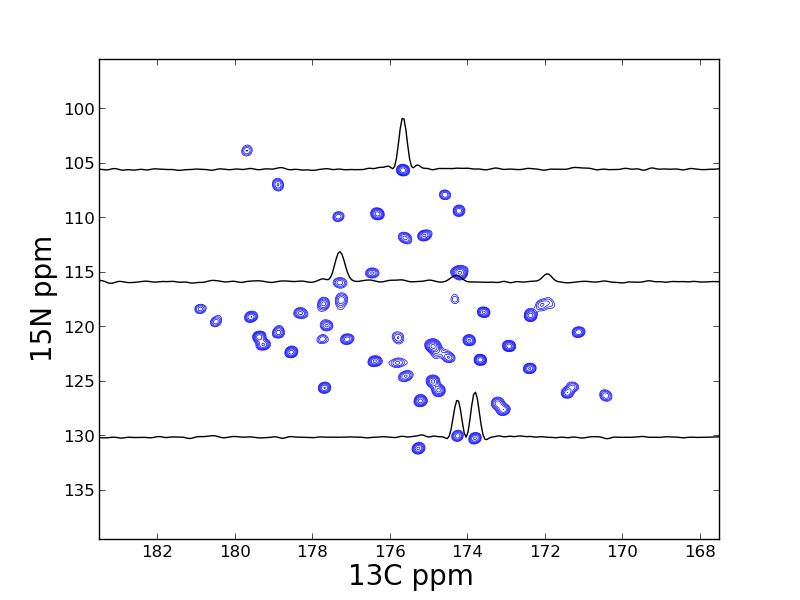
A contour plot of 2D data can created in a similar manner:
The plt.show() method raises an an interactive window for examining the plot:
Nmrglue Install
matplotlib can be used to create more complicated figures with annotations, ppmaxes and more. The Plotting Examples and Interactive Examplesshowcase some some of this functionality. For additional information see thematplotlib webpage
Additional resources¶
Detailed information about each module in nmrglue as well as the functionsprovided by that module can be found in the nmrglue Reference Guide orby using Python build in help system:
A number of Examples using nmrglue to interact withNMR data are available. Finally documentation for the following packagesmight be useful to users of nmrglue:
Google Colabs and NMRglue¶
Here is the code that has been used in colabs …
Nmr Glue
Once the software has been installed, the tutorial is downloaded and
unpacked
putting us in a position to follow the tutorial.
For example,
Nmrglue Bruker
and so on.
This notebook demonstrates automatic phase correction algorithms implemented for nmrglue. Two standard algorithms are implemented:
ACME algorithm by Chen Li et al. Journal of Magnetic Resonance 158 (2002) 164-168
Naive peak minima minimisation
The outputs for the two algorithms are shown below. Automatic phase correction can be used through the addition of an autops function to the proc_base set alongside the algorithm name to employ for scoring of phase. Custom algorithms can be provided via the same parameter.
